Post-transcriptional gene expression control is ubiquitous. It connects the transcriptional input from genes to the translational output in the form of proteins, in the end leading to the manifestation of phenotypes and adaptation.
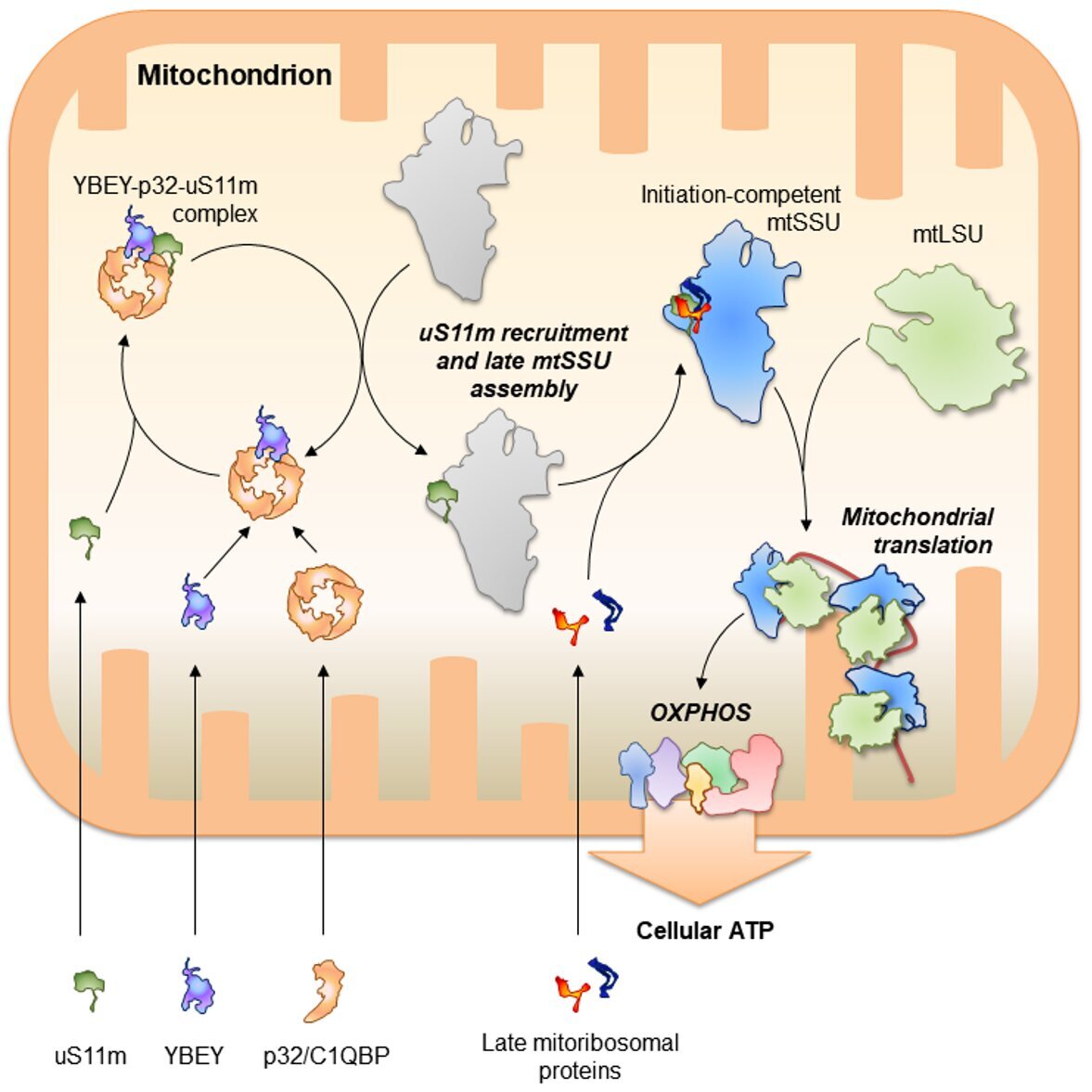
Our research is focused on post-transcriptional mechanisms used by relatively simple bacterial-type genetic systems (bacteria and their distant descendants, mitochondria) to globally shape - and eventually adjust - their gene expression.
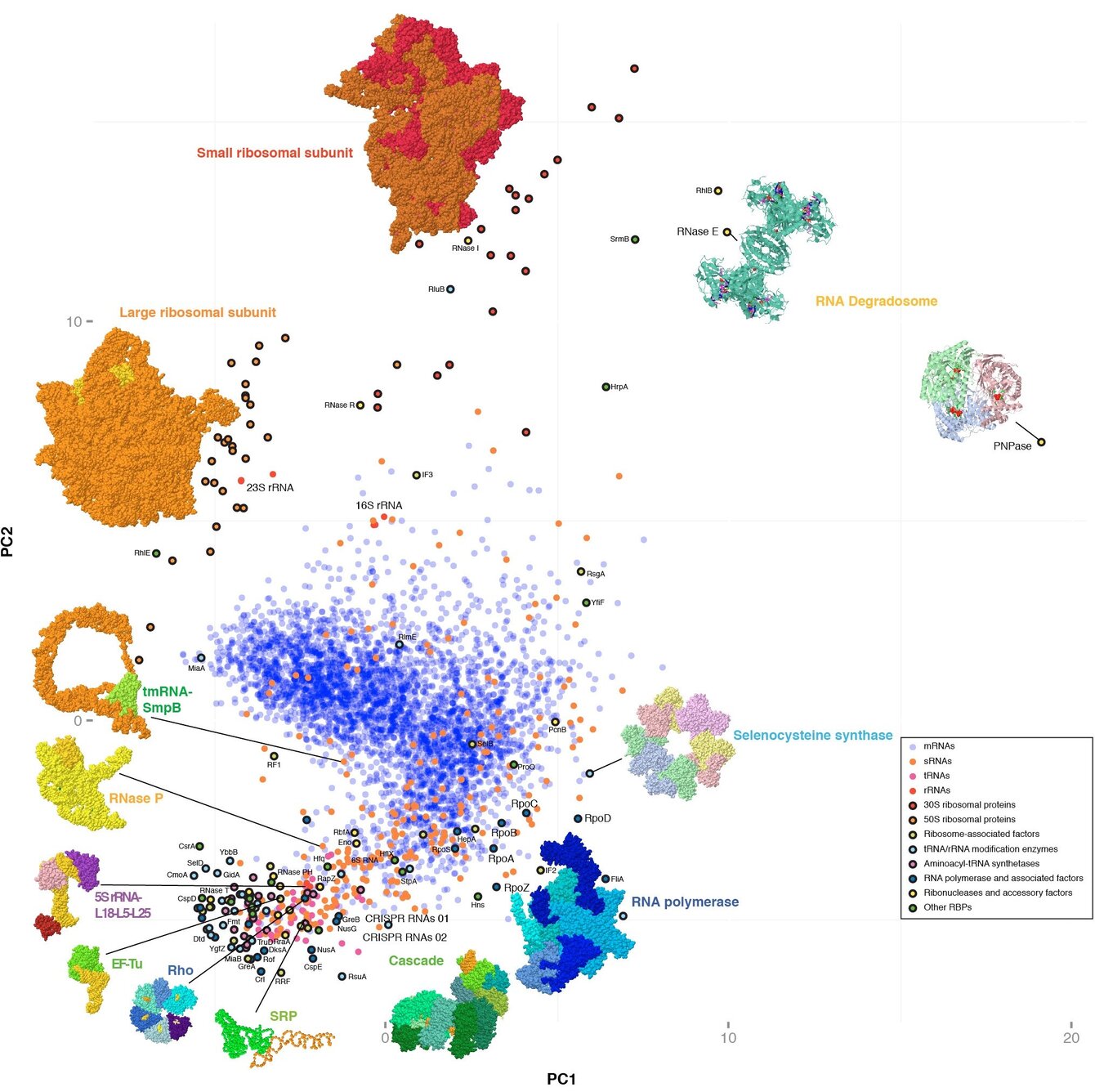
We are particularly interested in biology of highly conserved pleiotropic RNA-binding proteins (RBPs), noncoding RNAs, and the ribonucleoprotein particles (RNPs) they make up together.
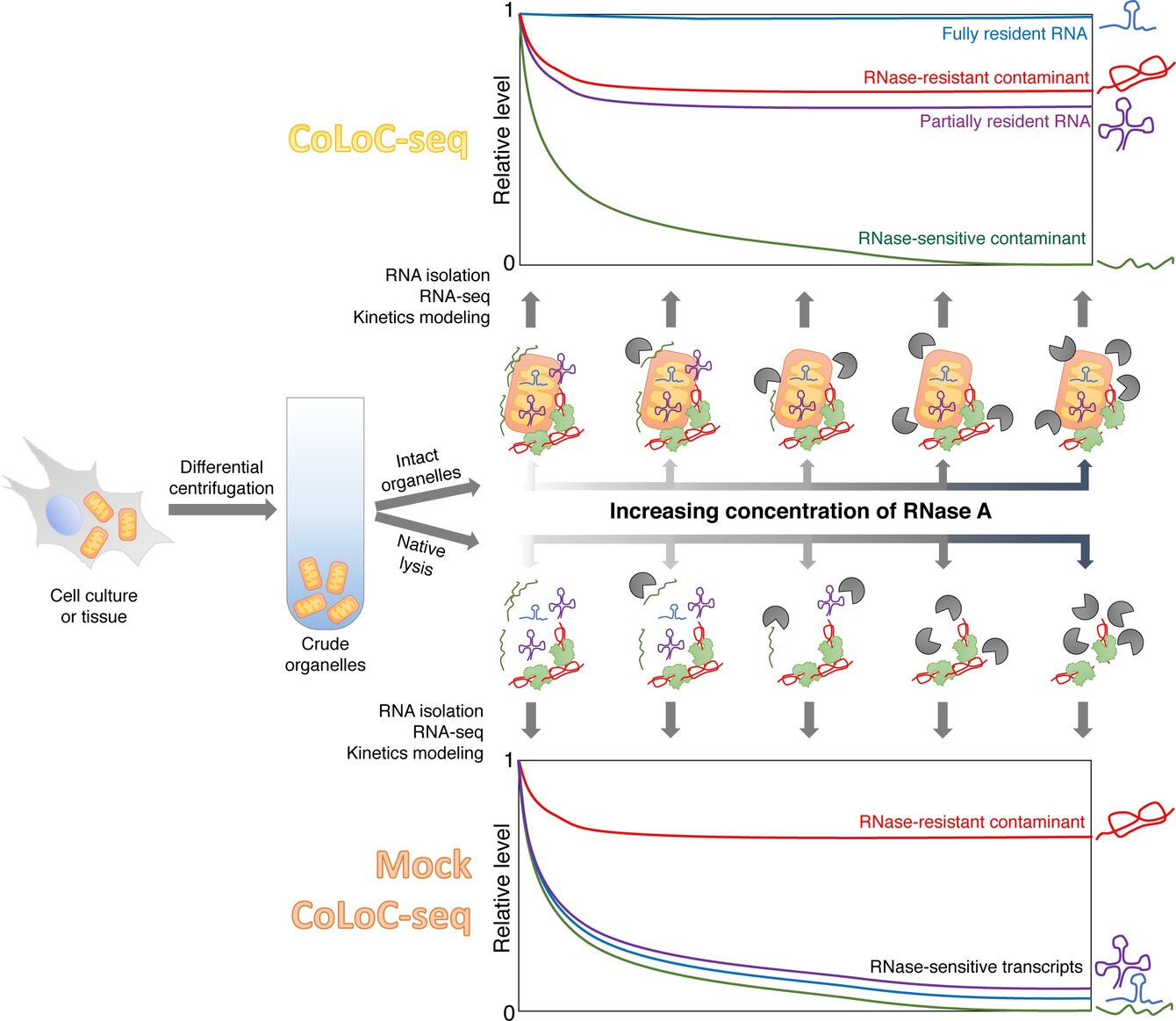
By using and developing a wide variety of methods – from biochemistry to systems-level approaches and experimental evolution – we aim at understanding how these fascinating regulators work at all levels (from elementary molecular acts to their physiological consequences) and why they are so deeply conserved across large phylogenetic clades. We are also interested in theoretical aspects of RNA-binding hubs.