Mitochondrial aminoacyl-tRNA synthetases in healthy and under disease conditions
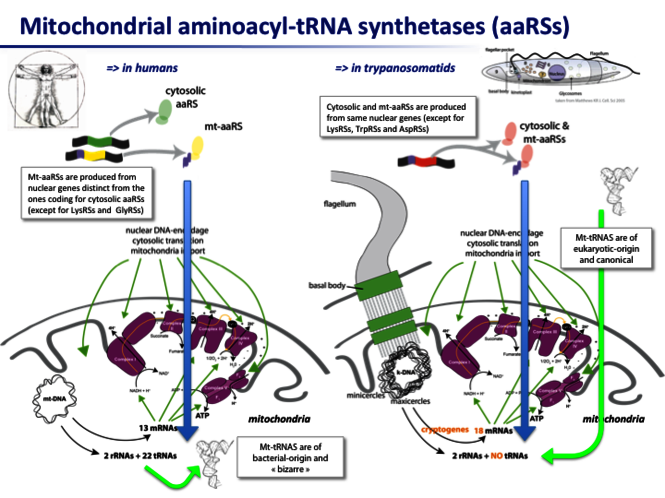

Aminoacyl-tRNA synthetases (aaRSs), key enzymes in mRNA translation process, catalyze the specific attachment of each amino acid to the cognate tRNA(s). With few exceptions, there are 20 aaRSs in each translation machinery of any organism. While mitochondrial (mt)-tRNAs are transcribed from the mt-DNA in humans, they are all imported from the nucleus into trypanosomatids. All mt-aaRSs are encoded by the nucleus, but from genes that are either mostly distinct in humans or mostly common in trypanosomatids from those coding for the cytosolic aaRSs. Mt-addressed proteins have a mitochondrial targeting sequence (MTS), most frequently at the N-terminus and cleaved after import by a process poorly defined but essential for protein's delivery and functionality. Mt-aaRSs remain insufficiently characterized despite i) the abundance of descriptions of patients mutated in their mt-aaRS-encoding genes, with poorly understood organs-specific clinical manifestations; and (ii) the presence of specific sequence insertions in aaRSs from trypanosomatids, suggesting unique properties, making them potential targets for the fight against related-infections. We propose that some mt-aaRSs possess multiple crucial role/peculiarities in different tissues and organisms, besides their known canonical function of tRNA aminoacylation. These will be searched for using complementary approaches and investigations in human and trypanosomatid cellular models.
