Targeted deletions in human mitochondrial DNA engineered by Type V CRISPR–Cas12a system
Natalia Nikitchina 1 , Anne-Marie Heckel 1 , Nikita Shebanov 1 , Ilya Mazunin 2 , 3 , Ivan Tarassov 1 , * ,Nina Entelis 1 , *
1 UMR7156—Molecular Genetics, Genomics, Microbiology, CNRS / University of Strasbourg, Strasbourg 67000, France 2 Department of Biology and Genetics, Petrovsky Medical University, Moscow 119435, Russia 3 Research Centre for Medical Genetics, Moscow 115478, Russia
NAR Molecular Medicine, Volume 2, Issue 2, April 2025, ugaf021, https://doi.org/10.1093/narmme/ugaf021
JOURNAL ARTICLE EDITOR'S CHOICE
Abstract
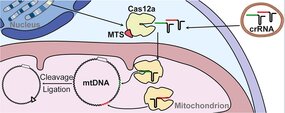
Mutations in mitochondrial DNA (mtDNA) contribute to various neuromuscular diseases, with severity depending on heteroplasmy level when mutant and wild-type mtDNA coexist within the same cell. Developing methods to model mtDNA dysfunction is crucial for experimental therapies. Here, we adapted the Type V CRISPR–AsCas12a system, which recognizes AT-rich PAM sequences, for targeted editing of human mtDNA. We demonstrated that mitochondrial targeting sequence (MTS) from Neurospora crassa ATPase subunit 9 efficiently addressed the AsCas12a effector nuclease into human mitochondria. When programmed with two CRISPR RNAs (crRNAs) targeting distant regions of mtDNA, the mito-AsCas12a can cleave mtDNA, enabling generation of deletions in cultured human cells. Next generation sequencing of the deletions boundaries confirmed mtDNA ligation after the cleavage by the mitoCRISPR–AsCas12a system. Therefore, we provide experimental data proving that a CRISPR system has potential to be used for precise mtDNA manipulation, offering a promising tool for generating predefined deletions in mtDNA and creating cellular models of mitochondrial disorders.